BisQue Software
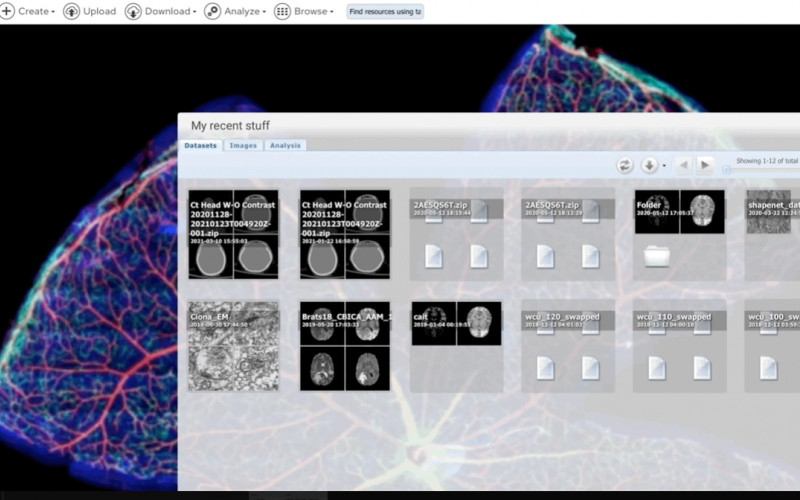
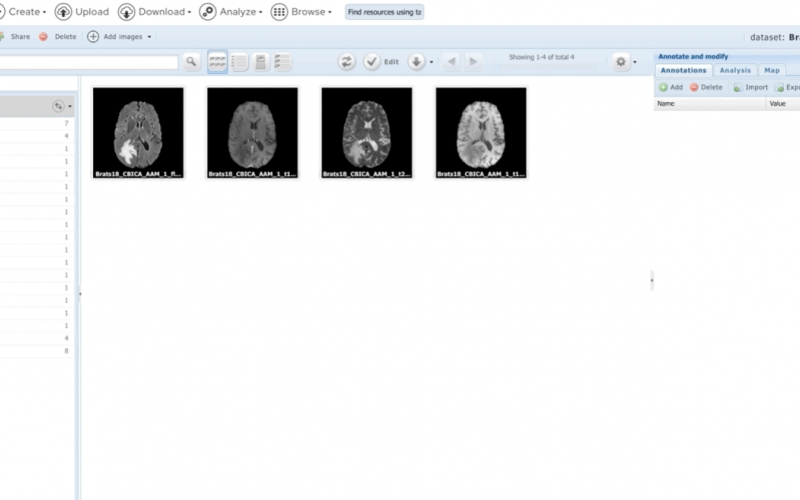
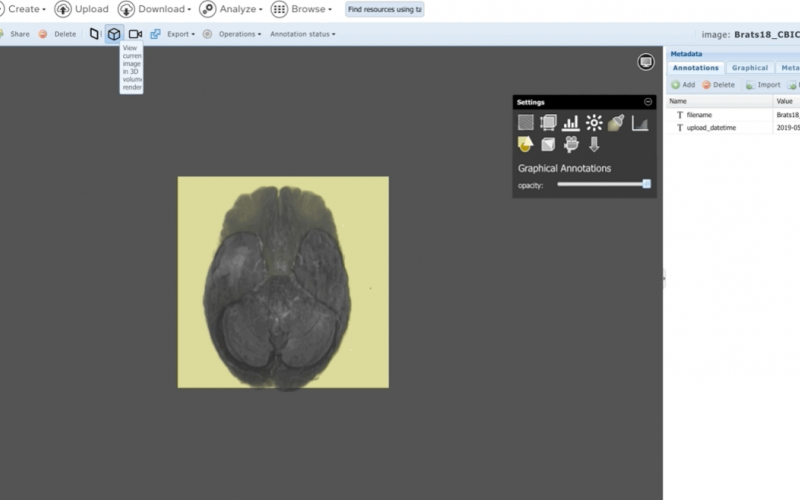
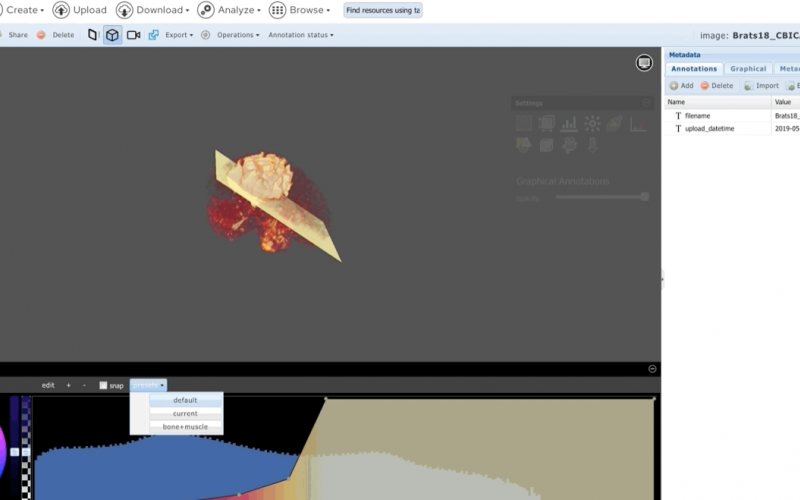
Bisque (Bio-Image Semantic Query User Environment) : Store, visualize, organize and analyze images in the cloud. Bisque was developed for the exchange and exploration of biological images. The Bisque system supports several areas useful for imaging researchers from image capture to image analsysis and querying. The bisque system is centered around a database of images and metadata. Search and comparison of datasets by image data and content is supported. Novel semantic analyses are integrated into the system allowing high level semantic queries and comparison of image content.
Overview
- Free and open-source
- Flexible textual and graphical annotations
- Cloud scalability: PBs of images, millions of annotations
- Distributed storage: local, iRODS, S3
- Integrated image analysis, high-throughput with Condor
- Analysis in MATLAB, Python, Java+ImageJ
- 100+ biological image formats
- Very large 5D images (100+ GB)
Documentation
The official documentation covers the BisQue cloud service running live at UCSB, module development for the platform, and the BQAPI. If you have any questions, feel free to reach out. We will be continuously updating the documentation so check back often for updates!
Source Code
https://github.com/UCSB-VRL/bisqueUCSB
Related Papers
- 2019: Latypov, M.I., Khan, A., Lang, C.A. et al. Integr Mater Manuf Innov 8: 52. https://doi.org/10.1007/s40192-019-00128-5
- 2019: Polonsky, A.T., Lang, C.A., Kvilekval, K.G. et al. Integr Mater Manuf Innov 8: 37. https://doi.org/10.1007/s40192-019-00126-7
- 2018: Trigkakis D., Todorovic S., Preece J., Meier A., Elser J., Jaiswal P., Kvilekval K., Fedorov D., Manjunath B.S. https://bigdatahealth.ucsb.edu/sites/default/files/publications/qt26h4h1c5.pdf
- 2017: D.V. Fedorov, K.G. Kvilekval, B.S. Manjunath, B. Doheny, S. Sampson, R.J. Miller https://bigdatahealth.ucsb.edu/sites/default/files/publications/dlfa.pdf
- 2016: D. Fedorov, R.J. Miller, K. Kvilekval, B. Doheny, S. Sampson, B.S. Manjunath https://escholarship.org/uc/item/0315h984
- 2016: J. Preece, J. Elser, P. Jaiswal, K. Kvilekval, D. Fedorov, B.S. Manjunath, R. Kitchen, X. Xu, D. Trigkakis, S. Todorovic, S. Carbon https://bigdatahealth.ucsb.edu/sites/default/files/publications/qt2xm742zt.pdf
- 2010: Kvilekval K, Fedorov D, Obara B, Singh A, Manjunath BS.;26(4):544-52. https://doi.org/10.1093/bioinformatics/btp699. Epub 2009 Dec 22. PMID: 20031971.
Contact
Amil Khan: amil [at] ucsb [dot] edu
Connor Levenson: clevenson [at] ucsb [dot] edu
Chandrakanth Gudavalli: chandrakanth [at] ucsb [dot] edu